Observing Mitosis with Fluorescence Microscopy
Interphase
Between mitotic divisions, a normal resting or actively growing cell exists in a state known as interphase, in which the chromatin forms a highly diffuse, fibrous network that is being continuously transcribed by enzymes within the nucleus. Before the cell enters the mitosis sequence, it first undergoes a DNA synthesis (or S) phase where each chromosome is duplicated to produce an identical pair of sister chromatids joined together by a specific DNA sequence known as a centromere. Centromeres are crucial to segregation of the daughter chromatids during mitosis.
View a second, third, and fourth fluorescence image of interphase.
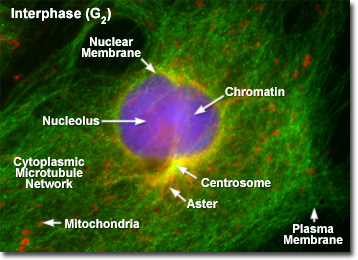
Illustrated in the fluorescence digital image above is a rat kangaroo (PtK2) kidney cell in the second gap (G(2)) period of interphase. The chromatin is stained with a blue fluorescent probe (DAPI), while the microtubule network is stained green (Alexa Fluor 488) and cellular mitochondria are stained with a red dye (MitoTracker Red CMXRos). The centrosomes act as the principal organizing center for the cell and function to nucleate a cytoplasmic array of microtubules, which project outward towards the plasma membrane. During interphase, the centrioles and other components of the centrosomal matrix are duplicated, but remain bound into a single unit on one side of the nucleus, as presented in the fluorescence micrograph. At the onset of mitosis, the centrosome complex divides with each centriole pair becoming the center of an organized network of radial microtubules.
When observed through the microscope, the cell nucleus is well defined and surrounded by the nuclear envelope (or membrane) during interphase. Within the nuclear membrane are one or more nucleoli, which appear as spherical, dense structures when stained with fluorescent or absorbing dyes. On the outer periphery of the nucleus are two important organelles termed centrosomes, which were formed by duplication of a single copy during the last division cycle. Each centrosome contains a pair of centrioles that extrude visible microtubule networks through the cytoplasm in radial arrays, known as asters (derived from the term stars). Even though the chromosomes have been duplicated during the DNA synthesis (S) phase, individual chromatids are not visible in late interphase because the chromosomes still exist in the form of loosely packed chromatin fibers.
In higher eukaryotic cells, the non-membranous centrosome (also commonly referred to as the microtubule-organizing center) functions through the cell cycle to organize the intracellular network of spindle and cytoskeletal microtubules. The pair of centrioles located at the center of the centrosome is not essential for cell division. In fact, most plants lack centrioles, and the organelles can be eliminated in animal cells through laser microsurgery with no apparent effect on spindle formation during mitosis.
Although very little activity is observable in the cell nucleus with fluorescence microscopy during interphase and the period is not considered to be a formal step in mitosis, this stage represents an essential preparation for cell division because the chromosomes are replicated during interphase. In addition to the synthesis phase, which occurs during the central portion of interphase, the cell cycle also consists of two gap (abbreviated G) stages that precede and follow the synthesis phase. For most animal cells, the interphase portion accounts for approximately 90 percent of the cell cycle, whereas mitosis is accomplished in the remaining period.
The first gap period is referred to as G(1) and starts shortly after mitosis, but prior to the synthesis (S) phase. The second gap period (G(2)) follows the synthesis phase and occurs just before mitosis. These gap periods exhibit intensive biochemical activity resulting in a doubling of the cell size with a similar increase in critical enzymes, ribosomes, mitochondria, carbohydrates, structural proteins, lipids, and other biomolecules and organelles that will be needed as the cell prepares for division. Collectively, the G and S periods are known as interphase.
The timeframe for completion of the entire cell cycle (the gap, synthesis, mitosis, and cytokinesis phases) varies depending upon the organism. For example, the common bean cell completes the cycle in about 19 hours, with 7 hours dedicated to the S period and two approximately equal length gap phases of 5 hours each. In this species, mitosis lasts about two hours. In contrast, mouse fibroblast cells in culture feature a cycle that lasts 22 hours, with mitosis being completed in only one hour. The synthesis phase in these mammalian rodent cells takes almost 10 hours, while G(1) lasts almost as long, or about 9 hours. The second gap phase (G(2)) is about two hours long.
When cells are forced to stop growing due to lack of nutrients or contact inhibition, they inevitably stop in the first gap (G(1)) phase. From this observation, it appears that essential biochemicals are synthesized during the G(1) period that either inhibit or stimulate the S phase and the rest of the cell cycle, thus determining whether or not division will take place.
BACK TO MITOSIS WITH FLUORESCENCE MICROSCOPY