Fluorescence Microscopy and Live-Cell Imaging
Fluorescent Proteins
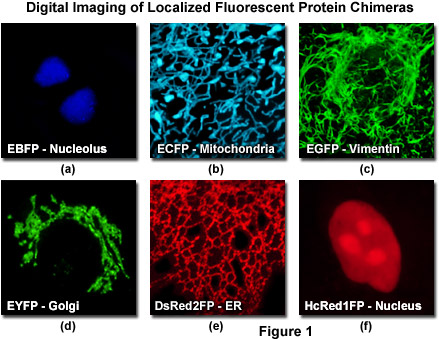
The discovery and development, over the past decade, of fluorescent proteins from a wide variety of marine organisms has initiated a revolution in the study of cell behavior by providing convenient markers for gene expression and protein targeting in living cells and organisms. The most widely used of these fluorescent proteins, the green fluorescent protein (GFP) first isolated from the jellyfish Aequorea victoria, can be attached to virtually any protein of interest and still fold into a fluorescent molecule. The resulting GFP fusion product can be used to localize previously uncharacterized proteins or to visualize and track known proteins to further understand cellular events. The use of fluorescent proteins as a minimally invasive tool for studying protein dynamics and function has been stimulated by the engineering of genetic variants with improved brightness, photostability and expression properties (see Figure 1). Cells that express gene products tagged with fluorescent proteins can be imaged with low light intensities over many hours to provide useful information about changes in the steady-state distribution of a protein over time.
Introduction to Fluorescent Proteins - The discovery of green fluorescent protein in the early 1960s ultimately heralded a new era in cell biology by enabling investigators to apply molecular cloning methods, fusing the fluorophore moiety to a wide variety of protein and enzyme targets, in order to monitor cellular processes in living systems using optical microscopy and related methodology. When coupled to recent technical advances in widefield fluorescence and confocal microscopy, including ultrafast low light level digital cameras and multitracking laser control systems, the green fluorescent protein and its color-shifted genetic derivatives have demonstrated invaluable service in many thousands of live-cell imaging experiments.
The Fluorescent Protein Color Palette - A broad range of fluorescent protein genetic variants have been developed over the past several years that feature fluorescence emission spectral profiles spanning almost the entire visible light spectrum. Extensive mutagenesis efforts in the original jellyfish protein have resulted in new fluorescent probes that range in color from blue to yellow and are some of the most widely used in vivo reporter molecules in biological research. Longer wavelength fluorescent proteins, emitting in the orange and red spectral regions, have been developed from the marine anemone Discosoma striata and reef corals belonging to the class Anthozoa. Still other species have been mined to produce similar proteins having cyan, green, yellow, orange, red, and far-red fluorescence emission. Developmental research efforts are ongoing to improve the brightness and stability of fluorescent proteins, thus improving their overall usefulness.
Fluorescent Proteins Derived from Aequorea victoria - Just in the past decade we have witnessed a truly remarkable expansion in the palette of Aequorea-based fluorescent proteins, largely driven by the innovative studies from Roger Tsien's laboratory. We now have jellyfish proteins that span an 80-nanometer portion visible spectrum from deep blue to yellow-green, providing a wide choice of genetically encoded markers for studies in cell biology. Most of the fluorescent proteins that are commonly used today have been modified through mutagenesis to optimize their expression in biological systems. Continued efforts using directed evolution approaches will no doubt improve the spectral characteristics, photostability, maturation time, brightness, acid resistance, and utility of the fluorescent protein tags for cellular imaging.
Anthozoa Fluorescent Proteins - Although promising candidates are now available in every Anthozoa fluorescent protein spectral class, in most cases there remains no EGFP equivalent, in terms of photostability and other critical areas of performance (with the exception of pH stability). New additions to the blue and cyan region feature substantially improved brightness and photostability, and any of the orange fluorescent proteins are excellent choices for long-term multicolor imaging. In addition, although brighter than EGFP, photostability is still suboptimal for the yellow fluorescent proteins, whereas the red and far-red variants are among the dimmest in all spectral classes. Further compounding the problem is the potential for aggregation artifacts due to poorly folding proteins, regardless of the spectral class or supposedly monomeric characteristics. Given that most of these proteins have only been introduced in the past couple of years, we remain optimistic that in the future, bright and photostable additions will become available for all spectral classes.
Imaging Parameters for Fluorescent Proteins - The wide spectrum of fluorescent proteins and derivatives uncovered thus far are quite versatile and have been successfully employed in almost every biological discipline from microbiology to systems physiology. These unique probes have proven extremely useful as reporters for gene expression studies in both cultured cells and entire animals. In living cells, fluorescent proteins are most commonly utilized to track the localization and dynamics of proteins, organelles, and other cellular compartments, as well as a tracer of intracellular protein trafficking. Quantitative imaging of fluorescent proteins is readily accomplished with a variety of techniques, including widefield, confocal, and multiphoton microscopy, to provide a unique window for exposing the intricacies of cellular structure and function.
Optical Highlighter Fluorescent Proteins - Protein chromophores that can be activated to initiate fluorescence emission from a quiescent state (a process known as photoactivation), or are capable of being optically converted from one fluorescence emission bandwidth to another (photoconversion), represent perhaps the most promising approach to the in vivo investigation of protein lifetimes, transport, and turnover rates. Appropriately termed molecular or optical highlighters, photoactivated fluorescent proteins generally display little or no initial fluorescence under excitation at the imaging wavelength, but dramatically increase their fluorescence intensity after activation by irradiation at a different (usually lower) wavelength. Photoconversion optical highlighters, on the other hand, undergo a change in the fluorescence emission bandwidth profile upon optically-induced changes to the chromophore. These effects result in the direct and controlled highlighting of distinct molecular pools within the cell.
Practical Considerations for using Fluorescent Proteins - Fluorescent proteins are quite versatile imaging probes and have been successfully employed in virtually every biological discipline ranging from microbiology to systems physiology. These ubiquitous probes are extremely useful as reporters for gene expression studies in cultured cells, excised tissues, and whole animals. In live cells, fluorescent proteins are most commonly used to track the localization and dynamics of proteins, organelles, and other cellular compartments, while they can also be used to assess protein-protein interactions through the use of resonance energy transfer techniques (FRET). This review provides some general tips for the practical aspects of using and imaging enhanced green fluorescent protein (EGFP) and newer members of the color palette.
Interactive Java Tutorials
Choosing Filter Combinations for Fluorescent Proteins - Fluorescence filter combinations designed to image fluorescent proteins must be carefully chosen to maximize the level of emission intensity presented to the detector while simultaneously reducing the number of unwanted photons from autofluorescence or bleed-through by other fluorophores. The broad absorption and emission spectral profiles exhibited by most fluorescent proteins offer a wide range of choice in filters, which are usually optimized for use with a specific detection system (human eye, digital camera, or photomultiplier). This interactive tutorial is designed to enable the identification of critical filter parameters, including the center wavelength, bandwidth region, and dichromatic mirror cut-on wavelength, which are necessary for imaging fluorescent proteins.
Choosing Fluorescent Proteins for Dual Labeling Experiments - The broad excitation and emission spectral profiles exhibited by fluorescent proteins and their color-shifted genetic variants often require specialized considerations when designing live-cell imaging experiments using two or more of these unique probes simultaneously. Of primary concern are potential bleed-through artifacts resulting from the significant degree of emission spectral overlap usually exhibited by fluorescent protein combinations. This interactive tutorial explores matching fluorescent proteins for dual labeling investigations with regards to spectral bandwidth and overlap, excitation efficiency, emission window dimensions, and other parameters necessary to design logical experiments.
Fluorescence Resonance Energy Transfer with Fluorescent Proteins - Fluorescent proteins are increasingly being applied as non-invasive probes in living cells due to their ability to be genetically fused to proteins of interest for investigations of localization, transport, and dynamics. In addition, the spectral properties of fluorescent proteins are ideal for measuring the potential for intracellular molecular interactions using the technique of Förster (or fluorescence) resonance energy transfer (FRET) microscopy. Because energy transfer is limited to distances of less than 10 nanometers, the detection of FRET provides valuable information about the spatial relationships of fusion proteins on a sub-resolution scale. This interactive tutorial explores various combinations of fluorescent proteins as potential FRET partners and provides information about critical resonance energy transfer parameters, as well as suggestions for microscope optical filter and light source configuration.
Optical Highlighter Fluorescent Proteins - Photoactivated fluorescent proteins generally display little or no initial fluorescence under excitation at the imaging wavelength, but dramatically increase their fluorescence intensity after activation by irradiation at a different (usually lower) wavelength. Photoconversion optical highlighters, on the other hand, undergo a change in the fluorescence emission bandwidth profile upon optically-induced changes to the chromophore. This interactive tutorial explores the optical conversion of several useful highlighter probes and simulates how these proteins would be viewed in an actual confocal microscope.
Fluorescent Protein Fluorophore Maturation Mechanisms - Autocatalytic formation of the fluorophore (also referred to as a chromophore) within the shielded environment of the polypeptide backbone during fluorescent protein maturation follows a surprisingly unified mechanism, especially considering the diverse natural origins of these useful biological probes. Shortly after synthesis, most fluorescent proteins slowly mature through a multi-step process that consists of folding, initial fluorophore ring cyclization, and subsequent modifications of the fluorophore. The spectral properties of fluorescent proteins are dependent upon the structure of the fluorophore as well as the localized interactions of amino acid residues in the immediate vicinity, and in some cases, residues far removed from the fluorophore. The interactive tutorials in this section explore fluorophore formation in a wide variety of spectrally diverse fluorescent proteins deduced from crystallographic studies.
PA-GFP Chromophore Photoactivation - The highly unique photophysical properties of wild-type green fluorescent protein (wild-type GFP) were thoroughly investigated during the mid-1990s, and served as a foundation for the creation of the first useful optical highlighter designed specifically for photoactivation studies. Termed PA-GFP (for Photo Activatable Green Fluorescent Protein), this optical highlighter was developed by improving the photoconversion efficiency of the native chromophore from a predominately neutral form to a species that is anionic in character. By replacing the threonine at position 203 with a histidine residue (T203H) in wild-type GFP, researchers produced a variant having negligible absorbance in the region between 450 and 550 nanometers, thus dramatically enhancing contrast between the non-activated and activated species.
Dronpa Fluorescent Protein Chromophore Photoswitching - The most prominent and well-studied photoswitchable fluorescent protein is named Dronpa (named after a fusion of the Ninja term for vanishing and photoactivation), which is a monomeric variant derived from a stony coral tetramer. Dronpa fluorescent protein exhibits an absorption maximum at 503 nanometers (arising from the anionic, deprotonated chromophore) with a minor peak at 390 nanometers (from the neutral, protonated chromophore). The anionic chromophore emits green fluorescence with a maximum at 518 nanometers and has a brightness level almost 2.5 times that of EGFP. Dronpa photoswitching occurs in part by interconversion between the deprotonated (on state; bright) and protonated (off state; dark) forms. Illumination at 488 nanometers drives Dronpa to the dark species after which the fluorescent protein can be subsequently switched back on by brief illumination at 405 nanometers. This cycle can be repeated several hundred times without significant photobleaching.
Photoconversion of Kaede/Eos Highlighters - One of the most useful classes of optical highlighters encompasses the growing number of fluorescent proteins reported to undergo photoconversion from one emission wavelength to another. Unlike photoactivatable fluorescent proteins, these probes are readily tracked and imaged in their native emission state prior to photoconversion, making it easier to identify and select regions for optical highlighting. The first report of a photoconvertable highlighter was a tetrameric fluorescent protein isolated from the stony Open Brain coral, Trachyphyllia geoffroyi, which can be photoconverted from green to red fluorescence emission by illumination with ultraviolet light. The discovery of this highlighter was serendipitous, as are many important discoveries. It occurred when the researchers accidentally left a test tube containing the protein on a laboratory bench near a window, and then astutely observed the shift from green to red. The unusual color transition prompted investigators to name the protein Kaede, after the leaves of the Japanese maple tree that turn from green to red in the fall.
Excited-State Proton Transfer - Among the most unique features of wild-type green fluorescent protein (wt-GFP) is the fact that illumination with either ultraviolet or blue-cyan light gives rise to green fluorescence having a maximum wavelength at approximately 507 nanometers. The bimodal absorption spectrum of wt-GFP features a large peak at 395 nanometers (the A band) and a much smaller peak at 475 nanometers (the B band). The lesser B band corresponds to the anionic chromophore, which demonstrates normal photophysics, whereas the predominant A band corresponds to the neutral chromophore that would normally be expected to emit blue light (peaking at approximately 450 nanometers) upon excitation in the ultraviolet. However, when excited with light ranging from 370 to 400 nanometers, the tyrosine residue in the neutral chromophore of wt-GFP becomes a strong acid and transfers a proton through a novel hydrogen bond network to generate an excited state anion (a process known as excited-state proton transfer; ESPT). It is the anionic form of the chromophore that emits green light.
Literature Sources
General Fluorescent Protein References - The disciplines of cellular and molecular biology are being rapidly and dramatically transformed by the application of fluorescent proteins developed from marine organisms as fusion tags to track protein behavior in living cells. The most widely used of these probes, green fluorescent protein, can be attached to virtually any target of interest and still fold into a viable fluorescent species. The resulting chimera can be employed to localize previously uncharacterized proteins or to visualize and track known proteins to further understand critical events at the cellular and molecular levels. This section features a bibliography of literature sources for review articles and original research reports on the discovery, applications, and continued development of fluorescent proteins.
ZEISS Campus Fluorescent Protein Reference Library - Application of the growing class of fluorescent proteins capable of forming an intrinsic chromophore to the observation of living cells and animals has almost single-handedly launched and fueled a new era in biology and medicine. These powerful research tools have provided investigators with a mechanism of fusing a genetically encoded optical probe to a practically unlimited variety of protein targets in order to examine living systems using fluorescence microscopy and related technology. The references listed below point to review articles that should provide the starting point for a thorough understanding of fluorescent protein technology.
Biosensor Fluorescent Protein References - By directly coupling the advanced microscopic technique of fluorescence resonance energy transfer (FRET) to the ubiquitous green fluorescent protein and its multispectral variants fused with selected biopolymers, ingenious researchers have created a new class of biosensors capable of elucidating mechanisms of signaling, enzymatic cleavage, and other critical functions in living cells. This section features a bibliography of literature sources for review articles and original research reports on the construction, applications, and continued development of biosensor fluorescent proteins.
Optical Highlighter Fluorescent Protein References - The ability to selectively initiate or alter fluorescence emission profiles in photoconversion optical highlighter proteins renders these probes as excellent tools for exploring protein behavior in living cells. As the fluorescence intensity (or color spectrum) of highlighters occurs only after photon-mediated conversion, newly synthesized non-photoactivated protein pools remain unobserved and do not complicate experimental results. This section lists sources for review articles and original research reports on optical highlighter fluorescent proteins.
Fluorescent Protein Photobleaching References - The field of cell biology is rapidly being transformed by the application of fluorescent proteins as fusion tags to track dynamic behavior in living cells. In this regard, fluorescence recovery after photobleaching (FRAP) is often employed to selectively destroy fluorescent molecules within a region of interest with a high-intensity laser, followed by monitoring the recovery of new fluorescent molecules into the bleached area over a period of time with low-intensity laser light. The resulting information can be used to determine kinetic properties, including the diffusion coefficient, mobile fraction, and transport rate of the fluorescently labeled molecules. The selected references in this section point to important literature sources for information on FRAP with fluorescent proteins.
Fluorescent Protein FRET References - Understanding the dynamic interactions between proteins within living cells is fundamental to a basic knowledge of the underlying concepts that guide molecular and cellular biology. Over the past few years, the rapid development of fluorescent proteins and their application as fusion products and biosensors has significantly expanded the molecular toolkit available for probing the mysteries of cellular physiology and pathology. In this regard, fluorescence (or Förster) resonance energy transfer (FRET) is emerging as a powerful optical microscopy technique for examining physiological processes with high temporal and spatial resolution. The references listed in this section highlight important literature sources for review articles and original research reports on the construction and applications of fluorescent proteins for resonance energy transfer experiments.
Internet-Based Education on Fluorescent Proteins - Despite the explosive growth of the Internet (in terms of the World Wide Web) as an informational resource for the original scientific literature pertaining to fluorescent protein investigations, there remains an obvious void in educational Websites targeted at beginning students and novices in the field. To address this issue, educational sites dedicated to optical microscopy and digital imaging being constructed and hosted at The Florida State University are turning their attention to the increasing application of fluorescent proteins for live-cell imaging studies. The primary focus of this effort is to create new sections of the sites that address the structure and properties of fluorescent proteins as well as optimizing their utility in imaging experiments.
Internet Resources
Fluorescent Protein Vector Commercial Sources - A variety of fluorescent proteins, available as recombinant DNA plasmid vectors designed for transfection of mammalian cells or transformation of bacteria, are commercially available from a number of distributors. Most of the vectors containing fluorescent protein DNA sequences have been codon-optimized for expression in mammalian cells and contain antibiotic genes for selection of stable mutants having relatively constant expression levels. The vectors often contain multiple cloning sequences that enable researchers to easily insert their gene of interest for fusion to the fluorescent protein. Other common features in fluorescent protein vectors include a human cytomegalovirus (CMV) promoter, a Kozak translation initiation site, an early mRNA polyadenylation signal, and a bacterial antibiotic gene.
Live Cell Imaging - One of the foremost targets in the life sciences is to understand the structure, function, and behavior of living organisms, and with evolving advances in technology, such as the development of confocal microscopy and fluorescent probes, it has become possible to pursue this goal at the cellular and subcellular levels. Still, working with and imaging live cells can be a complex, if not daunting, task to microscopists unfamiliar with the techniques and tools that are available. The following is a compilation of resources that offer overviews, background information, interactive forums, frequently asked questions, protocols, and hints that should aid any microscopist attempting to enter into this important, burgeoning field.
Microscopy Specimen Chambers - The demands of modern confocal microscopy, especially those involving imaging of living cells and tissues, require that researchers take special precautions with their specimens. Indeed, simple microscope slides are unsuitable for many applications, resulting in the development of a broad range of specimen chambers, which can often supply the flexibility a microscopist needs. The list of resources in this section exemplifies the great variety of specimen chambers that are available today and should help visitors locate the products that are best suited for their specific scientific pursuits.
Fluorescence Filters - Fluorescence microscopy relies heavily on the ability to select a specific wavelength region for excitation of the specimen and gathering secondary emission during image formation. Recent advances in interference filter design have resulted in highly accurate filters that cover a wide range of bandpass profiles, ranging from just a few to tens and hundreds of nanometers. Listed below is a compilation of manufacturers that design, manufacture, and supply fluorescence filters to the microscopy community. The products that they offer will meet most system requirements, but if a specialized application necessitates a filter with an unusual spectral range or other unique specifications, many of the companies will fabricate custom filters.
Fluorescent Protein Principle Investigators - Many of the scientists involved with research targeting various aspects of cell biology are using fluorescent proteins as imaging probes for cell structure, function, and dynamics. Several of the principle investigators have built extensive websites detailing their laboratories, and these sites are quite useful to visitors interested in learning more about this exciting and rapidly evolving research arena. Included in the information on a majority of the websites linked below are the current research interests, curriculum vitas, publications, lists of laboratory personnel, contact information, educational tutorials, image galleries, and digital videos.
Contributing Authors
Jennifer Lippincott-Schwartz and George H. Patterson - Cell Biology and Metabolism Branch, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland, 20892.
David W. Piston - Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee, 37232.
Richard N. Day - Department of Cellular and Integrative Physiology, Indiana University School of Medicine, 635 Barnhill Dr., Indianapolis, Indiana, 46202.
Robert E. Campbell - Department of Chemistry, University of Alberta, Edmonton, Alberta, Canada, T6G2G2
Matthew J. Parry-Hill, Nathan S. Claxton and Michael W. Davidson - National High Magnetic Field Laboratory, 1800 East Paul Dirac Dr., The Florida State University, Tallahassee, Florida, 32310.
BACK TO FLUORESCENCE MICROSCOPY
