Fluorescence Digital Image Gallery
Indian Muntjac Deer Skin Fibroblast Cells
|
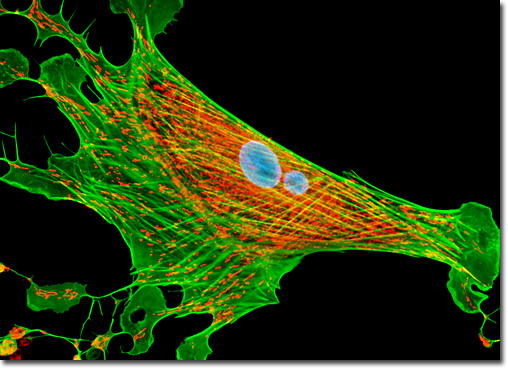
Over the past few years, the discovery and development of naturally occurring fluorescent proteins and mutated derivatives have rapidly advanced to center stage in the investigation of a wide spectrum of intracellular processes in living organisms. These biological probes have provided scientists with the ability to visualize, monitor, and track individual molecules with high spatial and temporal resolution in both steady-state and kinetic experiments. A variety of marine organisms have been the source of more than 25 fluorescent proteins and their analogs, which arm the investigator with a balanced palette of non-invasive biological probes for single, dual, and multispectral fluorescence analysis. Among the advantages of fluorescent proteins over the traditional organic and new semiconductor probes described above is their response to a wider variety of biological events and signals. Coupled with the ability to specifically target fluorescent probes in subcellular compartments, the extremely low or absent photodynamic toxicity, and the widespread compatibility with tissues and intact organisms, these biological macromolecules offer an exciting new frontier in live-cell imaging. An adherent culture of Indian Muntjac deer skin fibroblast cells (illustrated above) was transfected with a pDsRed-Mitochondria plasmid subcellular localization vector, thus localizing a red fluorescent protein tag to the intracellular mitochondrial network. The culture was subsequently fixed and labeled with Hoechst 33342 and Alexa Fluor 488 conjugated to phalloidin, targeting DNA and filamentous actin, respectively. Images were recorded in grayscale with a QImaging Retiga Fast-EXi camera system coupled to an Olympus BX-51 microscope equipped with bandpass emission fluorescence filter optical blocks provided by Omega Optical. During the processing stage, individual image channels were pseudocolored with RGB values corresponding to each of the fluorophore emission spectral profiles. |
© 1995-2025 by Michael W. Davidson and The Florida State University. All Rights Reserved. No images, graphics, software, scripts, or applets may be reproduced or used in any manner without permission from the copyright holders. Use of this website means you agree to all of the Legal Terms and Conditions set forth by the owners.
This website is maintained by our
|