Fluorescence Digital Image Gallery
Human Cervical Adenocarcinoma Cells (HeLa)
|
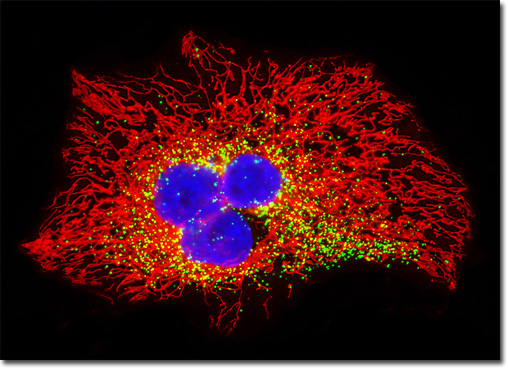
Mutagenesis experiments with green fluorescent protein have produced a large number of variants with improved folding and expression characteristics, which have eliminated wild-type dimerization artifacts and fine-tuned the absorption and fluorescence properties. One of the earliest variants, known as enhanced green fluorescence protein (EGFP), contains codon substitutions (commonly referred to as the S65T mutation) that alleviates the temperature sensitivity and increases the efficiency of GFP expression in mammalian cells. Proteins fused with EGFP can be observed at low light intensities for long time periods with minimal photobleaching. Enhanced green fluorescent protein fusion products are optimally excited by the 488-nanometer spectral line from argon and krypton-argon ion lasers in confocal and fluorescence microscopy. This provides an excellent biological probe and instrument combination for examining intracellular protein pathways along with the structural dynamics of organelles and the cytoskeleton. The HeLa carcinoma cell culture featured in the digital image above was transfected with an EGFP-peroxisomal targeting signal 1 (PTS1) fusion protein and stained with MitoTracker Red CMXRos. These fluorescent probes target the peroxisomes and intracellular microtubular network, respectively. The visible light absorption maximum of the EGFP-PTS1 chimera is 488 nanometers and the emission maximum occurs at 507 nanometers. In addition, the specimen was simultaneously stained with Hoechst 33342 (targeting the DNA in the nucleus; blue emission). Images were recorded in grayscale with a QImaging Retiga Fast-EXi camera system coupled to an Olympus BX-51 microscope equipped with bandpass emission fluorescence filter optical blocks provided by Omega Optical. During the processing stage, individual image channels were pseudocolored with RGB values corresponding to each of the fluorophore emission spectral profiles. |
© 1995-2025 by Michael W. Davidson and The Florida State University. All Rights Reserved. No images, graphics, software, scripts, or applets may be reproduced or used in any manner without permission from the copyright holders. Use of this website means you agree to all of the Legal Terms and Conditions set forth by the owners.
This website is maintained by our
|