Fluorescence Digital Image Gallery
Rat Jejunum Myenteric Plexus Enteroglial Cells (EGC/PK060399egfr Line)
Enteroglial cells (known by the acronym: EGC) are believed to be an important part of the enteric nervous system, but much about the function of these cells is still unknown. To help facilitate future studies of the cells, A. Ruhl and coworkers developed a new method in 2001 for isolating and purifying enteroglial cells from the myenteric plexus, a network of nerve fibers located in the intestinal wall.
The Ruhl method involved enzymatic dissociation of myenteric plexus samples, the purification of enteric glial cells via complement-mediated cytolysis of contaminating cells, and transformation by retroviral gene transfer. The group then characterized the resulting clones both immunohistochemically and by dot-blot analysis. As a result of their efforts, a number of transformed EGC lines that retain their glial and functional characteristics have been established.
The EGC/PK060399egfr line is one of the new enteroglial cell lines created by Ruhl and his collaborators. Derived from the myenteric plexus of a rat jejunum, the cells grow adherently in culture and exhibit glial morphology. The source of the cells was an adult male member of the species Rattus rattus (Sprague-Dawley strain). The cells exhibit vigorous glial fibrillary acidic protein (GFAP), S-100, and vimentin immunoreactivities. They do not, however, display Thy-1.1, desmin, smooth muscle alpha-actin, or C3 complement receptor immunoreactivity. The EGC/PK060399egfr line is useful for characterization studies of enteric glial cells and may eventually aid in the better understanding of various intestinal disorders that are currently inadequately understood, such as inflammatory bowel disease.
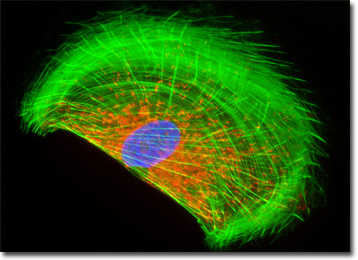
The mitochondria present in the culture of rat enteroglial cells featured in the digital image above were labeled with MitoTracker Red CMXRos, a derivative of X-rosamine. In addition, the culture was labeled for the cytoskeletal F-actin network and DNA in the cell nucleus with Alexa Fluor 488 conjugated to phalloidin and DAPI, respectively. Images were recorded with a QImaging Retiga Fast-EXi camera system coupled to an Olympus BX-51 microscope equipped with bandpass emission fluorescence filter optical blocks provided by Omega Optical.
Additional Fluorescence Images of Rat Enteroglial (EGC/PK060399egfr) Cells
Localizing the Golgi Network in EGC Enteroglial Cells - A culture of rat enteroglial cells was labeled with Oregon Green 488 conjugated to the lectin wheat germ agglutinin. Fluorescent wheat germ agglutinin conjugates are often used as probes for the Golgi network in mammalian cultures. The cells were also stained with Alexa Fluor 568 conjugated to phalloidin and DAPI, which target F-actin and DNA, respectively.
Targeting GFAP in Enteroglial Cells with Immunofluorescence - The adherent log phase culture of rat enteroglial cells presented in this section was immunofluorescently labeled with primary anti-glial fibrillary acidic protein (GFAP) mouse monoclonal antibodies followed by goat anti-mouse Fab fragments conjugated to Cy3. The culture was also stained for DNA with Hoechst 33258 and for F-actin with Alexa Fluor 488 conjugated to phalloidin.
Actin and Mitochondria Distribution in Rat Enteroglial Cells - The mitochondria and filamentous actin present in an EGC cell culture were targeted with MitoTracker Red CMXRos (red fluorescence) and Alexa Fluor 488 conjugated to phalloidin (green fluorescence), respectively. In addition, the culture was counterstained for DNA with DAPI.
Histone and Peroxisome Distribution in EGC Cell Cultures - In a double immunofluorescence experiment, an adherent monolayer culture of EGC cells was fixed, permeabilized, blocked with 10 percent normal goat serum, and treated with a cocktail of mouse anti-histones (pan) and rabbit anti-PMP 70 (peroxisomal membrane protein) primary antibodies, followed by goat anti-mouse and anti-rabbit secondary antibodies (IgG) conjugated to Texas Red and Alexa Fluor 488, respectively. The filamentous actin network was counterstained with Alexa Fluor 350 conjugated to phalloidin.
EGC Cell Cultures with Texas Red, Alexa Fluor 488, and DAPI - In this section, the featured culture of rat enteroglial cells was labeled with Texas Red conjugated to wheat germ agglutinin, a lectin that selectively binds with sialic acid residues. The culture was subsequently labeled for the cytoskeletal F-actin network with Alexa Fluor 488, and for DNA in the cell nucleus with the ultraviolet-absorbing probe DAPI.
Concanavalin A Localization of the Endoplasmic Reticulum in Rat Enteroglial Cells - A culture of EGC enteroglial cells was stained with Alexa Fluor 488 conjugated to the lectin concanavalin A, which selectively binds to alpha-mannopyranosyl and alpha-glucopyranosyl residues (primarily in the endoplasmic reticulum). Alexa Fluor 568 conjugated to phalloidin and DAPI were also used to label the culture, targeting filamentous actin and nuclear DNA, respectively.
Focal Adhesions in EGC Cell Cultures - The adherent culture of EGC cells featured in this section was immunofluorescently labeled with anti-vinculin mouse monoclonal primary antibodies followed by goat anti-mouse Fab heavy and light chain fragments conjugated to Rhodamine Red-X. In addition, the specimen was simultaneously stained for DNA with the ultraviolet-absorbing probe DAPI, and for the cytoskeletal filamentous actin network with Alexa Fluor 488 conjugated to phalloidin.
Visualizing the Actin Network, Mitochondria, and DNA Localization in Rat Enteroglial Cells - Using the popular combination of MitoTracker Red CMXRos, Alexa Fluor 488 conjugated to phalloidin, and DAPI, an adherent culture of EGC cells was first incubated with the MitoTracker, and then fixed, permeabilized and blocked with bovine serum albumen. The specimen was subsequently labeled with the phallotoxin conjugate, followed by the DAPI counterstain.
Distribution of Histones and the Golgi Complex in EGC Monolayer Cell Cultures - The proximity of the Golgi complex and nuclei in rat enteroglial cells was probed in a double immunofluorescence experiment with mouse anti-histones (pan) and rabbit anti-giantin primary antibodies. The antibody targets were visualized with goat secondary antibodies conjugated to Texas Red and Alexa Fluor 488, respectively, while the actin cytoskeletal framework was labeled with Alexa Fluor 350 conjugated to phalloidin.
Rat Enteroglial Cells (EGC) with Alexa Fluor 488, Alexa Fluor 568, and DAPI - In a manner similar to that employed for a specimen linked above, a culture of EGC enteroglial cells was stained with Alexa Fluor 488 conjugated to the lectin concanavalin A, which selectively binds to alpha-mannopyranosyl and alpha-glucopyranosyl residues (primarily in the endoplasmic reticulum). Alexa Fluor 568 conjugated to phalloidin and DAPI were also used to label the culture, targeting filamentous actin and nuclear DNA, respectively.
Visualizing the Microtubule and Actin Cytoskeletal Networks in EGC Cell Cultures - The rat enteroglial cells presented in this section were immunofluorescently labeled with mouse anti-alpha-tubulin primary antibodies followed by goat anti-mouse secondary antibodies conjugated to Alexa Fluor 568. In addition, the specimen was stained with Alexa Fluor 350 conjugated to phalloidin and SYTOX Green, targeting the filamentous actin network and nuclei, respectively.
BACK TO THE CULTURED CELLS FLUORESCENCE GALLERY
BACK TO THE FLUORESCENCE GALLERY
